González-Bello
Research themes
Antibacterials: Innovative approaches to face the superbug challenge. Unexplored targets. Drugs with novel mechanism of action. New diagnosis kits.
Main researcher(s)
Group members
| Maneiro Rey, María |
Postdoctoral Researcher |
|
| Canabal Facón, Rafael |
PhD Candidate |
|
| Casabella Amieiro, Braulio |
PhD Candidate |
|
| Outeiral Valiño, María Carmen |
PhD Candidate |
|
| Suárez de Cepeda Fuentes, Pilar |
PhD Candidate |
Research
Approaches to Unlock Bacterial Resistance
The current strategy for anti-infective drug discovery by the pharmaceutical industry has not changed significantly in the last 60 years. This mainly involves modifications of earlier classes that target a small number and the same type of bacterial functions, to make them more efficient and robust against resistant bacteria. However, resistance to these targets is widespread and well known. A new path in the antibiotic drug discovery approach is therefore urgently needed to control the increasing development and spread of SUPERBUGS, which are pathogens have even become resistant to multiple types of antibiotics. Our research group is focus on the development of novel antibacterial agents with an INNOVATIVE mechanism of action to treat deadly infections caused by multi-drug resistant Gram-negative pathogens, especially for priority bacteria as recently published by the WHO, i.e., Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacteriaceae. These pathogens are a major concern on patients with a weak immune system undergoing cancer chemotherapy, general surgery, organ transplant and dialysis, amongst others, for whom our ability to deal with secondary infections is crucial. We are also studying the potential of unexplored bacterial targets for the development of novel anti-tuberculosis agents, which is already considered as a top global health priority.
Three main strategies that deal with BACTERIAL VIABILITY, BACTERIAL RESISTANCE and BACTERIAL COMMUNICATION are currently explored in our research group.
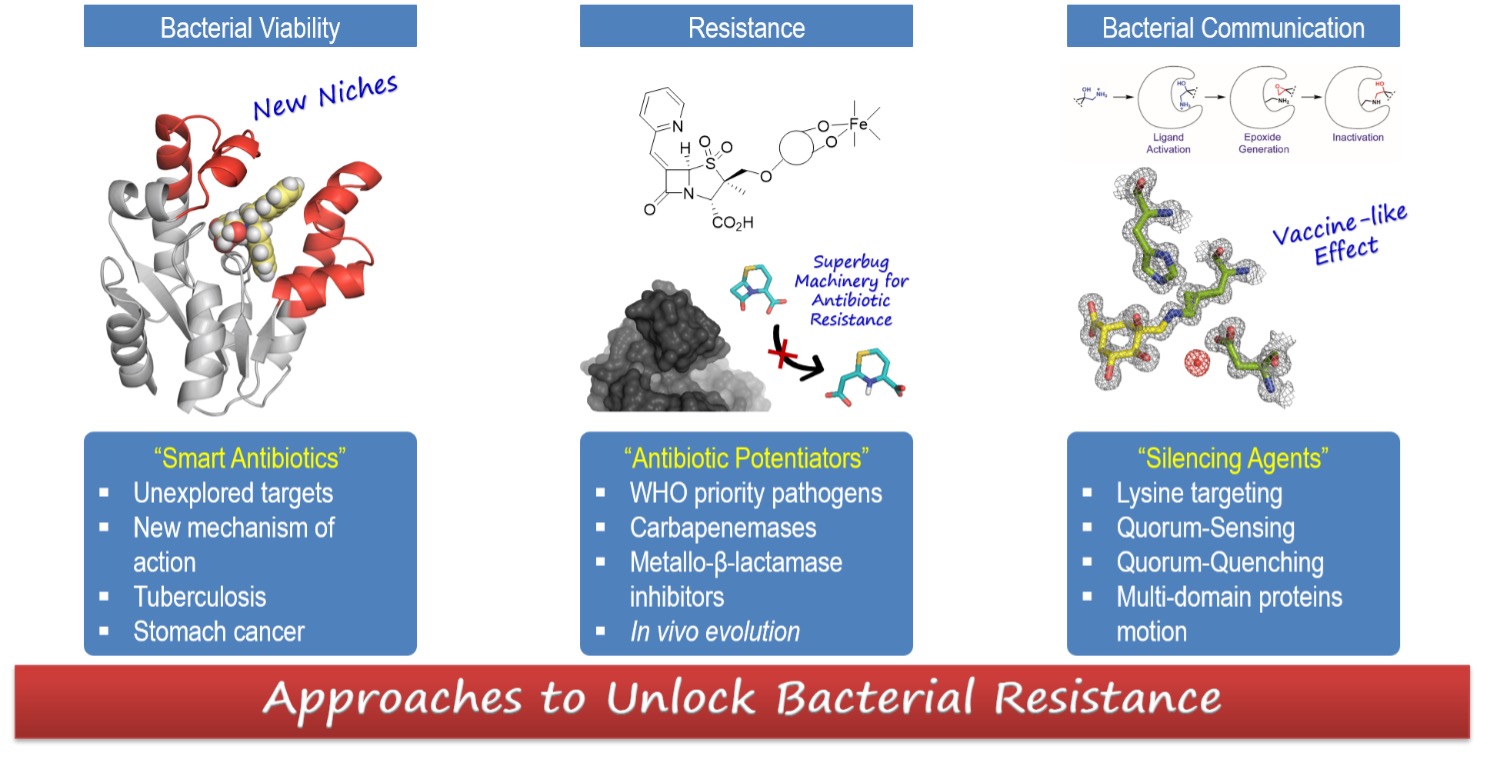
SMART ANTIBIOTICS – We are studying the therapeutic potential of several enzymes of the shikimic acid pathway (shikimate kinase, EPSP synthase) for antibiotic drug discovery. These unexplored targets are essential for the viability of the bacterium. Based on the essential enzyme motions for turnover (motion-based design), we have identified potent reversible competitive inhibitors that stabilize an inactive open conformation for catalysis (J. Med. Chem. 2016, 59, 5471, Chem. Eur. J. 2016, 22, 17988; Org. Chem. Front. 2019, 6, 2514). We have also developed inhibitors by rigidification of the high-energy active conformation that the enzyme recognizes for catalysis (J. Am. Chem. Soc. 2013, 135, 12366). We use a prodrug approach for internalization.
ANTIBIOTIC POTENTIATORS – We are working on the development of new antibiotic adjuvants that block the most relevant mechanism of bacterial resistance – the enzymatic inactivation of the antibiotic (β-lactams) by β-lactamases. From the drug discovery point of view, this combined drug therapy (antibiotic + antibiotic adjuvant) has the advantage that it is not necessary to expend effort in challenging and expensive identification of new targets. Specifically, we are focus on the most challenging β-lactamases, class D carbapenemases (OXA-23, OXA-24/40, OXA-48), metallo-β-lactamases and chromosomal class C enzymes from P. aeruginosa (AmpC), which are produced by the most dangerous superbugs in clinical settings. We have developed diverse penicillin-based sulfones that form stable indolizidine adducts with the enzyme that allows the in vitro and in vivo repurposing of carbapenem and cephalosphorin activity in those challenging bacterial strains (Antimicrob. Agents Chemother. 2017, 61, e01172; J. Antimicrob. Chemother. 2016, 7, 2171; Antimicrob. Agents Chemother. 2019, 63, e01092-19). We are also studying in the in vivo evolution of bacterial resistance, as an inspiring source for drug design (J. Antimicrob. Chemother. 2020, 75, 3209-3217; J. Antimicrob. Chemother. 2021, 76, 91-100).
SILENCING AGENTS – The attenuation of bacterial communication (Quorum-Quenching) makes the bacterium less able to establish successful infection and, in consequence, it is cleared by the host immune response. We are focus in disabling the Quorum Sensing (QS) transcriptional regulator LasR, which is the master regulator of QS in P. aeruginosa modulating the expression of more than 300 genes (ACS Nano 2015, 9, 5567). We are also interested in the design of smart irreversible compounds for the anti-virulence bacterial target type I dehydroquinase (DHQ1) enzyme, which acts as a virulence factor in vivo. We have developed diverse compounds non-bearing reactive electrophiles that cause the selective covalent modification of lysine residues (Org. Chem. Front. 2019, 6, 3127), and we have shown that DHQ1 catalyzes its self‐immolation by transforming an unreactive 2‐hydroxyethylammonium group into an epoxide that triggers the irreversible inhibition (J. Am. Chem. Soc. 2015, 137, 9333; Chem. Eur. J. 2020, 26, 8035).
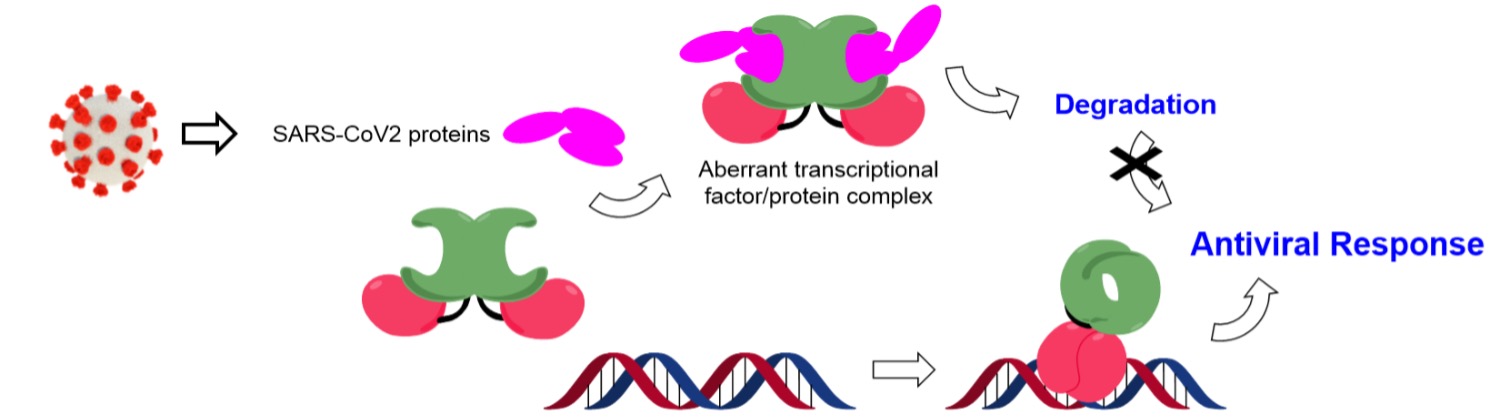
Approaches to Cancel the Immuno-Evasion and Suppression Mechanisms of SARS-CoV-2
The COVID-19 disease pandemic caused by the 2019 novel coronavirus infection (SARS-CoV-2) has become a global public health emergency, whose study is also the focus of our research. SARS-CoV-2 infection causes the disorder of natural and adaptive immunity that leads to tissue damage and systemic inflammation, which is the main cause of death of patients with this disease. Viral proteins often play a fundamental role in interfering with the host's immune response, however the molecular mechanism underlying the modulation of immune signaling pathways by SARS-CoV-2 is currently unknown. We became recently interested in exploring the molecular mechanism underlying this complex and challenging viral process, as well as in the development of small chemical entities able to modulate it. We are focused on the study in atomic detail of the mechanism of activation of the interferon signaling pathway that establish an antiviral state, which seems to be modulated by the expression of small proteins capable of inactivating the interferon production in the early stages of infection. In fact, the systematic removal of the interferon modulatory functions of the virus is a very promising approach for vaccine development.
Tools for Design
Our research group has specialized in the application of powerful complementary tools for drug design and discovery, such as protein X-ray crystallography and high-level computational studies that considers the dynamics and the mechanism of the biomacromolecule, thus, Molecular Dynamics simulations, Quantum Mechanics/Molecular Mechanics (QM/MM) studies. This allows us to approach more complex projects from a multidisciplinary perspective beyond chemistry, as well as to get a deeper understanding of the key factors for ligand binding, inhibition, or activity modulation. We have solved more than 30 crystal structures of diverse enzyme/ligand complexes and enzyme/ligand adducts, such as shikimate kinase from M. tuberculosis, type II dehydroquinase from M. tuberculosis and H. pylori and type I dehydroquinase from S. typhi and S. aureus. Based on the essential enzyme motion, we have also developed potent reversible competitive inhibitors with good in vitro and in vivo activity against H. pylori (Chem. Eur. J. 2016, 22, 17988). We have employed QM/MM simulations to determine the catalytic mechanism of the type II dehydroquinase (DHQ2) enzyme, a recognized target for antibiotic discovery, and to identify the determinants of the catalytic activity differences between DHQ2 homologous enzymes (Org. Biomol. Chem. 2018, 16, 4443), as well as the selective covalent modification mechanism of enzymes/proteins, either induced by ligand recognition (Chem. Eur. J. 2020, 26, 8035; Org. Chem. Front. 2019, 6, 3127), or by generation of reactive species upon irradiation (J. Org. Chem. 2018, 83, 13019; Chem. Sci. 2017, 8, 2621; Org. Chem. Front. 2019, 6, 99; Chem. Eur. J. 2020, 26, 15922;).




